ICCK Transactions on Emerging Topics in Artificial Intelligence
ISSN: 3068-6652 (Online)
Email: [email protected]


 Submit Manuscript
Edit a Special Issue
Submit Manuscript
Edit a Special Issue
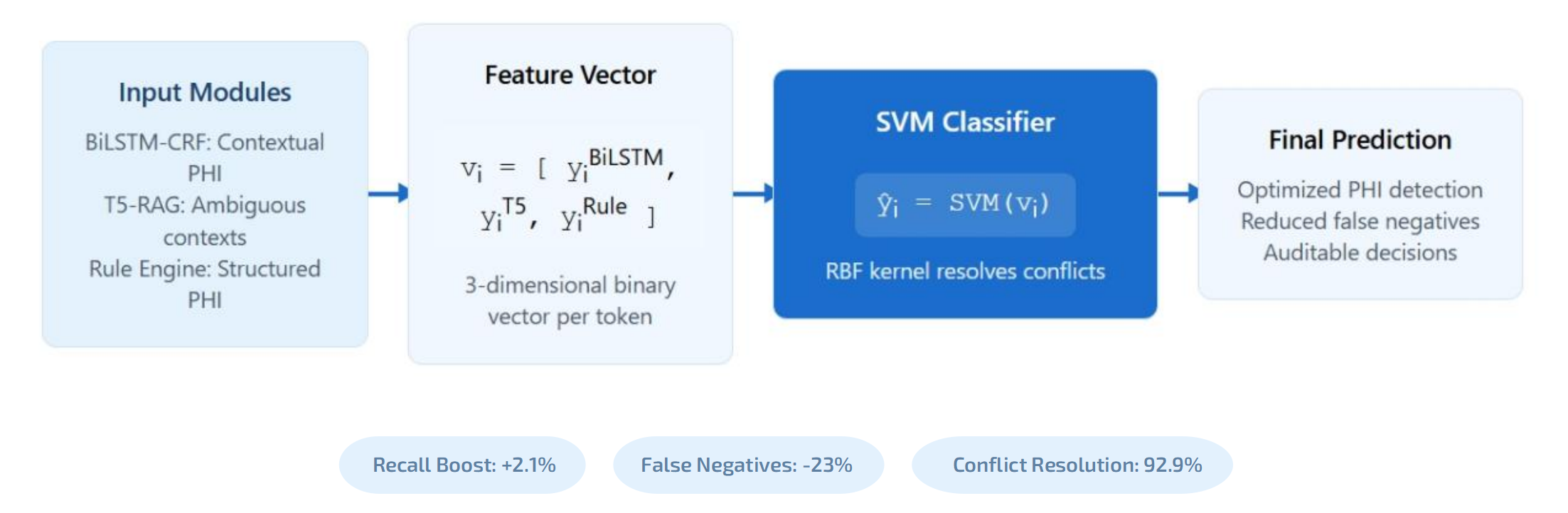
TY - JOUR AU - Ye, Kai PY - 2025 DA - 2025/11/12 TI - Hybrid Large Language Model and Rule-Based Framework for Automated PHI De-Identification in Clinical Notes JO - ICCK Transactions on Emerging Topics in Artificial Intelligence T2 - ICCK Transactions on Emerging Topics in Artificial Intelligence JF - ICCK Transactions on Emerging Topics in Artificial Intelligence VL - 3 IS - 1 SP - 1 EP - 8 DO - 10.62762/TETAI.2025.518010 UR - https://www.icck.org/article/abs/TETAI.2025.518010 KW - PHI de-identification KW - clinical NLP KW - large language models KW - hybrid systems KW - parameter-efficient fine-tuning (PEFT) KW - electronic health records KW - privacy preservation KW - retrieval-augmented generation (RAG) KW - rule-based NLP KW - biomedical text processing AB - The growing demand for secondary use of electronic health records (EHRs) in clinical research has amplified the importance of effective de-identification of protected health information (PHI) to comply with privacy regulations such as HIPAA. Manual annotation remains error-prone, time-consuming, and inconsistent across healthcare institutions, while existing automated systems often face trade-offs between accuracy, interpretability, and computational cost. This study proposes a novel hybrid de-identification framework that integrates neural, statistical, and rule-based approaches to achieve high recall, operational efficiency, and deployment feasibility in real-world healthcare settings. SN - 3068-6652 PB - Institute of Central Computation and Knowledge LA - English ER -
@article{Ye2025Hybrid,
author = {Kai Ye},
title = {Hybrid Large Language Model and Rule-Based Framework for Automated PHI De-Identification in Clinical Notes},
journal = {ICCK Transactions on Emerging Topics in Artificial Intelligence},
year = {2025},
volume = {3},
number = {1},
pages = {1-8},
doi = {10.62762/TETAI.2025.518010},
url = {https://www.icck.org/article/abs/TETAI.2025.518010},
abstract = {The growing demand for secondary use of electronic health records (EHRs) in clinical research has amplified the importance of effective de-identification of protected health information (PHI) to comply with privacy regulations such as HIPAA. Manual annotation remains error-prone, time-consuming, and inconsistent across healthcare institutions, while existing automated systems often face trade-offs between accuracy, interpretability, and computational cost. This study proposes a novel hybrid de-identification framework that integrates neural, statistical, and rule-based approaches to achieve high recall, operational efficiency, and deployment feasibility in real-world healthcare settings.},
keywords = {PHI de-identification, clinical NLP, large language models, hybrid systems, parameter-efficient fine-tuning (PEFT), electronic health records, privacy preservation, retrieval-augmented generation (RAG), rule-based NLP, biomedical text processing},
issn = {3068-6652},
publisher = {Institute of Central Computation and Knowledge}
}
 Copyright © 2025 by the Author(s). Published by Institute of Central Computation and Knowledge. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
Copyright © 2025 by the Author(s). Published by Institute of Central Computation and Knowledge. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. ICCK Transactions on Emerging Topics in Artificial Intelligence
ISSN: 3068-6652 (Online)
Email: [email protected]

Portico
All published articles are preserved here permanently:
https://www.portico.org/publishers/icck/