Abstract
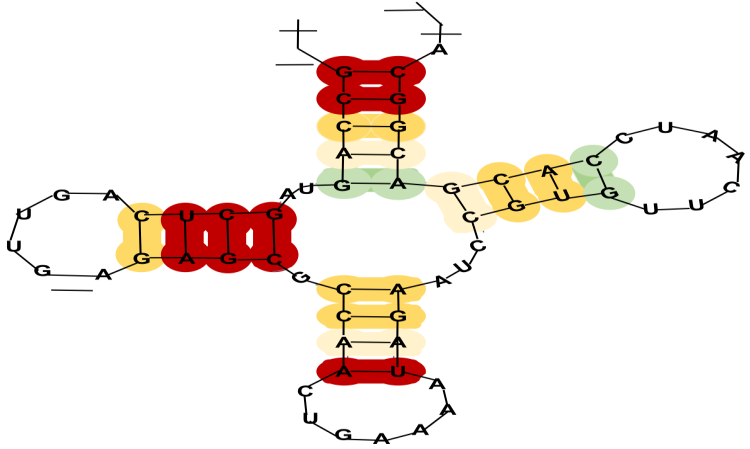
Long non-coding RNAs (lncRNAs) are important for plant growth, how plants respond to stress, and their overall development. However, it can be difficult to identify them accurately because they come in many structures and can look similar to coding RNAs. In this study, we introduce DeepPInc V2.0, a new tool that uses deep learning to analyze both the sequence and the secondary structure of RNAs, combining them in a DenseNet-CNN hybrid model. DeepPInc V2.0 outperforms existing tools on various plant datasets, achieving an accuracy of 94.2%, an F1-score of 0.93, and a Matthews Correlation Coefficient (MCC) of 0.88. It consistently outperforms seven leading tools in this area. Importantly, the model still works well even with incomplete or shortened sequences, which is a common issue in transcriptome studies. When we applied DeepPInc V2.0 to the wheat (Triticum aestivum) transcriptome under heat stress, we found over 27,000 possible lncRNAs. Of these, 1,830 were expressed differently, suggesting they may play a role in helping plants adapt to stress. These results demonstrate that DeepPInc V2.0 is a reliable and accurate platform for identifying lncRNAs in plants, facilitating the study of large RNA collections and understanding the functions of non-coding elements.
Data Availability Statement
Data will be made available on request.
Funding
This work was supported without any funding.
Conflicts of Interest
The authors declare no conflicts of interest.
Ethical Approval and Consent to Participate
Not applicable.
Cite This Article
APA Style
Pandey, S., & Sharma, R. (2025). A Deep Learning Approach for Long Non-coding RNA Identification in Plants: DeepPlnc V2.0. Biomedical Informatics and Smart Healthcare, 1(2), 79–88. https://doi.org/10.62762/BISH.2025.421075
Publisher's Note
ICCK stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and Permissions

Copyright © 2025 by the Author(s). Published by Institute of Central Computation and Knowledge. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (
https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.


 Submit Manuscript
Edit a Special Issue
Submit Manuscript
Edit a Special Issue
 Copyright © 2025 by the Author(s). Published by Institute of Central Computation and Knowledge. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
Copyright © 2025 by the Author(s). Published by Institute of Central Computation and Knowledge. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. 