Chinese Journal of Information Fusion
ISSN: 2998-3371 (Online) | ISSN: 2998-3363 (Print)
Email: [email protected]
 Submit Manuscript
Edit a Special Issue
Submit Manuscript
Edit a Special Issue
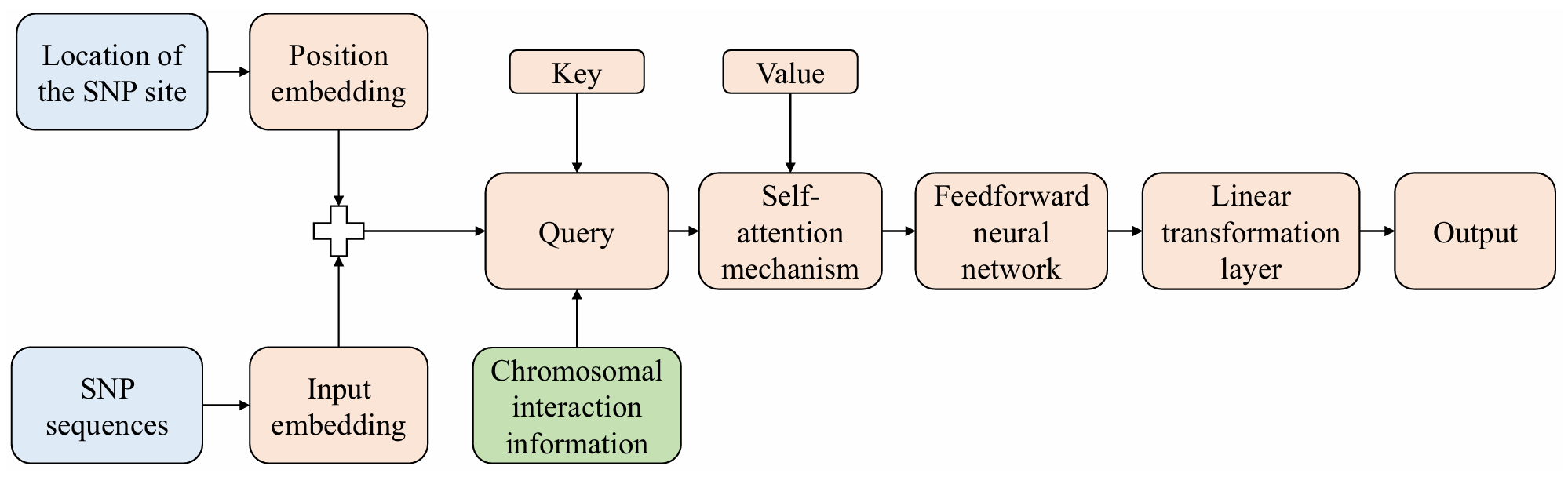
TY - JOUR AU - Chen, Ailing AU - Zou, Qingke AU - Yang, Xidi AU - Zhou, Jie PY - 2026 DA - 2026/02/08 TI - Transformer Fusing Chromosome Conformation and Genomic Information for Soybean Trait Prediction JO - Chinese Journal of Information Fusion T2 - Chinese Journal of Information Fusion JF - Chinese Journal of Information Fusion VL - 3 IS - 1 SP - 31 EP - 45 DO - 10.62762/CJIF.2025.226807 UR - https://www.icck.org/article/abs/CJIF.2025.226807 KW - transformer KW - information fusion KW - chromosome interaction KW - genomic prediction KW - soybean traits AB - Genomic information is increasingly leveraged for the precise prediction of crop traits, with the adoption of advanced genomic prediction techniques resulting in substantial improvements in both crop yield and quality. However, traditional genomic prediction methods exhibit notable limitations in capturing long-range dependencies and fully utilizing prior information from chromosome structure. In this work, two novel Transformer models fusing chromosome conformation and genomic information have been proposed. One is the chromosomal self-attention fusion model, which captures cross-chromosomal interactions more precisely by introducing chromosomal conformation information into the self-attention mechanism of the Transformer. The other is the chromatin interaction squeeze excitation model, which extracts global features of the chromosome from all single nucleotide polymorphism sites on each chromosome. It then employs the chromatin interaction matrix to perform a weighted fusion of these global features, enabling the effective utilization of inter-chromosomal information. In addition, two novel metrics are introduced to comprehensively assess the effectiveness of the internal self-attention mechanism. They quantify the concentration of attention while measuring the alignment between the attention distribution and the chromosomal interaction priors. Experiments show that the two proposed models exhibit significant advantages in predicting soybean oil content and protein. SN - 2998-3371 PB - Institute of Central Computation and Knowledge LA - English ER -
@article{Chen2026Transforme,
author = {Ailing Chen and Qingke Zou and Xidi Yang and Jie Zhou},
title = {Transformer Fusing Chromosome Conformation and Genomic Information for Soybean Trait Prediction},
journal = {Chinese Journal of Information Fusion},
year = {2026},
volume = {3},
number = {1},
pages = {31-45},
doi = {10.62762/CJIF.2025.226807},
url = {https://www.icck.org/article/abs/CJIF.2025.226807},
abstract = {Genomic information is increasingly leveraged for the precise prediction of crop traits, with the adoption of advanced genomic prediction techniques resulting in substantial improvements in both crop yield and quality. However, traditional genomic prediction methods exhibit notable limitations in capturing long-range dependencies and fully utilizing prior information from chromosome structure. In this work, two novel Transformer models fusing chromosome conformation and genomic information have been proposed. One is the chromosomal self-attention fusion model, which captures cross-chromosomal interactions more precisely by introducing chromosomal conformation information into the self-attention mechanism of the Transformer. The other is the chromatin interaction squeeze excitation model, which extracts global features of the chromosome from all single nucleotide polymorphism sites on each chromosome. It then employs the chromatin interaction matrix to perform a weighted fusion of these global features, enabling the effective utilization of inter-chromosomal information. In addition, two novel metrics are introduced to comprehensively assess the effectiveness of the internal self-attention mechanism. They quantify the concentration of attention while measuring the alignment between the attention distribution and the chromosomal interaction priors. Experiments show that the two proposed models exhibit significant advantages in predicting soybean oil content and protein.},
keywords = {transformer, information fusion, chromosome interaction, genomic prediction, soybean traits},
issn = {2998-3371},
publisher = {Institute of Central Computation and Knowledge}
}
 Copyright © 2026 by the Author(s). Published by Institute of Central Computation and Knowledge. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
Copyright © 2026 by the Author(s). Published by Institute of Central Computation and Knowledge. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. Chinese Journal of Information Fusion
ISSN: 2998-3371 (Online) | ISSN: 2998-3363 (Print)
Email: [email protected]

Portico
All published articles are preserved here permanently:
https://www.portico.org/publishers/icck/