Plant Innovation Journal
ISSN: pending (Online)
Email: [email protected]
 Submit Manuscript
Edit a Special Issue
Submit Manuscript
Edit a Special Issue
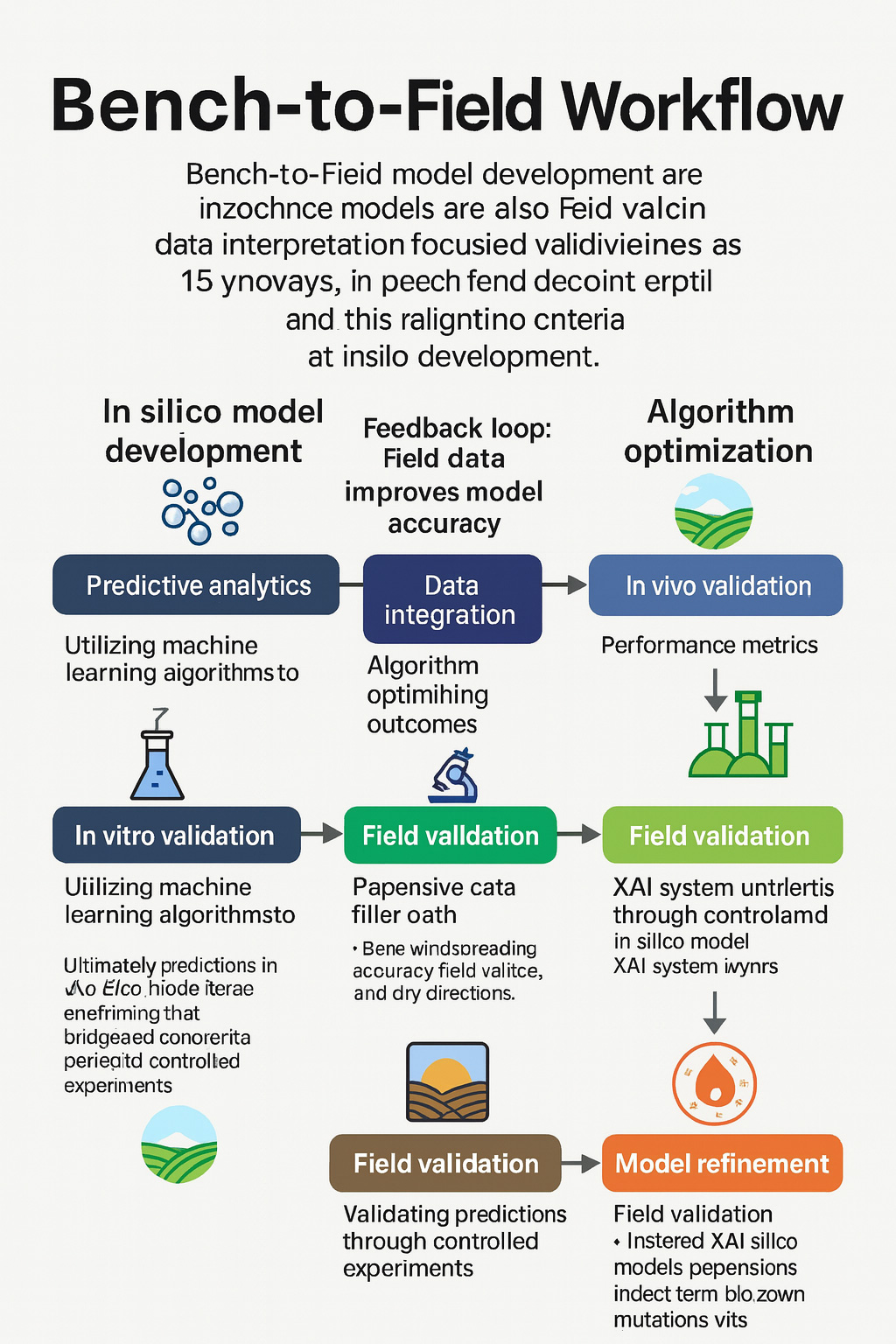
TY - JOUR AU - Rahimi, Mehdi PY - 2026 DA - 2026/01/04 TI - Bridging the Translational Divide: A Vision for AI-Guided Genome Editing from Bench to Field in Cereal Crops JO - Plant Innovation Journal T2 - Plant Innovation Journal JF - Plant Innovation Journal VL - 1 IS - 1 SP - 8 EP - 17 DO - 10.62762/PIJ.2025.121730 UR - https://www.icck.org/article/abs/PIJ.2025.121730 KW - CRISPR genome editing KW - cereal breeding KW - explainable artificial intelligence (XAI) KW - genotype-by-environment (G×E) AB - The mix of AI and CRISPR gene editing is changing how we upgrade grain crops, which feed much of the world. In this inaugural perspective, we propose a transformative framework to close the gap between computational prediction and field performance. Rather than presenting new data, we call for a paradigm shift toward explainable AI, digital twins, federated learning, and breeder-centric platforms. We argue that only through integrated, transparent, and collaborative systems can we realize the full promise of precision breeding for global food security. Still, translating computational predictions into successful crop performance in the field often fails or exhibits rapid performance decline. Here’s A critical analysis of the primary failure points - finding targets, making edits and growing plants, then testing them in different fields - and proposes a practical framework that fits the tricky biology of grains such as wheat, corn, and rice. We pull together realistic standards for picking models (focusing on transparent AI instead of hidden algorithms, testing under unexpected stresses), handling edits safely (methods for complex, repeated DNA patterns, designing with cell structure in mind, checking thoroughly for unintended changes), plus getting trials set up right (solid checks of genes meeting environments, repeating tests across locations, planning seed production to field timing smartly). We lay out a step-by-step process using clear AI to spot key traits, link gene edits with safety checks for side effects, while running virtual tests to predict how crops perform in different climates and genetics. Moving beyond conceptual proposals, we define a set of verifiable metrics to track progress from lab work to real-world use. Turning scattered tips into fixed rules helps avoid costly mistakes later, speeds up creating tough grain varieties, and makes results easier to verify worldwide. This piece speaks directly to those wanting practical steps matched to actual farming needs, answering calls for fresh approaches in crop research. SN - pending PB - Institute of Central Computation and Knowledge LA - English ER -
@article{Rahimi2026Bridging,
author = {Mehdi Rahimi},
title = {Bridging the Translational Divide: A Vision for AI-Guided Genome Editing from Bench to Field in Cereal Crops},
journal = {Plant Innovation Journal},
year = {2026},
volume = {1},
number = {1},
pages = {8-17},
doi = {10.62762/PIJ.2025.121730},
url = {https://www.icck.org/article/abs/PIJ.2025.121730},
abstract = {The mix of AI and CRISPR gene editing is changing how we upgrade grain crops, which feed much of the world. In this inaugural perspective, we propose a transformative framework to close the gap between computational prediction and field performance. Rather than presenting new data, we call for a paradigm shift toward explainable AI, digital twins, federated learning, and breeder-centric platforms. We argue that only through integrated, transparent, and collaborative systems can we realize the full promise of precision breeding for global food security. Still, translating computational predictions into successful crop performance in the field often fails or exhibits rapid performance decline. Here’s A critical analysis of the primary failure points - finding targets, making edits and growing plants, then testing them in different fields - and proposes a practical framework that fits the tricky biology of grains such as wheat, corn, and rice. We pull together realistic standards for picking models (focusing on transparent AI instead of hidden algorithms, testing under unexpected stresses), handling edits safely (methods for complex, repeated DNA patterns, designing with cell structure in mind, checking thoroughly for unintended changes), plus getting trials set up right (solid checks of genes meeting environments, repeating tests across locations, planning seed production to field timing smartly). We lay out a step-by-step process using clear AI to spot key traits, link gene edits with safety checks for side effects, while running virtual tests to predict how crops perform in different climates and genetics. Moving beyond conceptual proposals, we define a set of verifiable metrics to track progress from lab work to real-world use. Turning scattered tips into fixed rules helps avoid costly mistakes later, speeds up creating tough grain varieties, and makes results easier to verify worldwide. This piece speaks directly to those wanting practical steps matched to actual farming needs, answering calls for fresh approaches in crop research.},
keywords = {CRISPR genome editing, cereal breeding, explainable artificial intelligence (XAI), genotype-by-environment (G×E)},
issn = {pending},
publisher = {Institute of Central Computation and Knowledge}
}
 Copyright © 2026 by the Author(s). Published by Institute of Central Computation and Knowledge. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
Copyright © 2026 by the Author(s). Published by Institute of Central Computation and Knowledge. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. 
Portico
All published articles are preserved here permanently:
https://www.portico.org/publishers/icck/