Journal of Mathematics and Interdisciplinary Applications
ISSN: 3070-393X (Online)
Email: [email protected]
 Submit Manuscript
Edit a Special Issue
Submit Manuscript
Edit a Special Issue
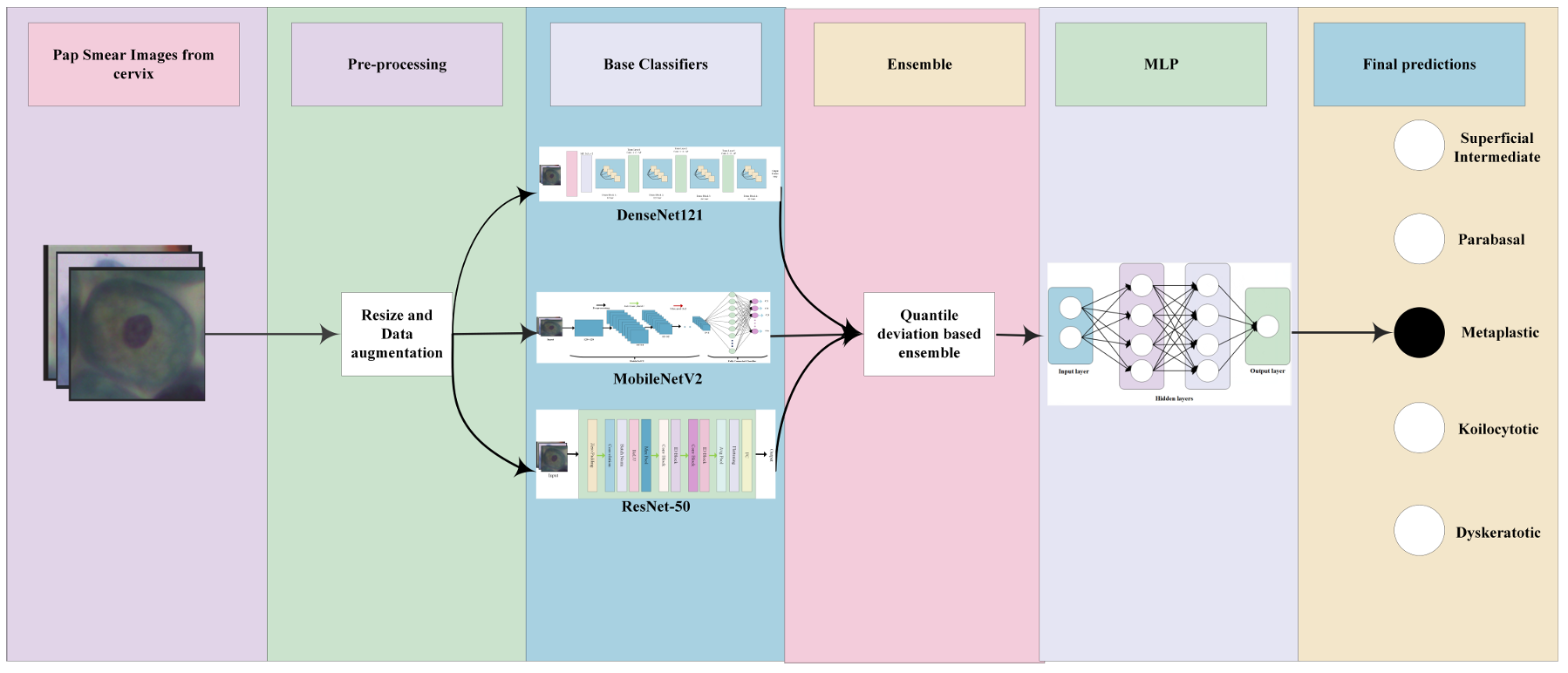
TY - JOUR AU - Hu, Hui AU - Huang, Jiewu AU - Cao, Shoumei AU - Dai, Anna PY - 2026 DA - 2026/02/07 TI - Quantile Deviation Ensemble Based on Multi-Layer Perceptrons for Cervical Cancer Classification with Uncertainty Perception JO - Journal of Mathematics and Interdisciplinary Applications T2 - Journal of Mathematics and Interdisciplinary Applications JF - Journal of Mathematics and Interdisciplinary Applications VL - 2 IS - 1 SP - 12 EP - 27 DO - 10.62762/JMIA.2026.403659 UR - https://www.icck.org/article/abs/JMIA.2026.403659 KW - cervical cytology classification KW - uncertainty quantification KW - ensemble learning KW - quantile deviation KW - deep learning AB - The accurate classification of cervical cytology in Pap smear images remains a critical challenge in computer-aided diagnosis, largely due to the inherent uncertainty and subtle morphological variations among different pathological categories. To address this, we propose a novel uncertainty-aware ensemble framework that integrates statistical quantile analysis with deep learning for robust and interpretable classification. Our framework first leverages three deep convolutional neural networks DenseNet121, MobileNetV2, and ResNet-50 as base feature extractors. Instead of employing naive ensemble strategies, we introduce a quantile deviation based weighting mechanism to dynamically assess and integrate the prediction confidence of each model, explicitly quantifying performance bias across different probability quantiles. This approach not only enhances ensemble stability but also provides a statistical measure of model uncertainty. Subsequently, the weighted probabilistic outputs are fed into a multi-layer perceptron (MLP) for further non-linear optimization and decision refinement, forming a hybrid statistical deep learning pipeline. Evaluated on the publicly available SIPaKMeD dataset, our framework achieves an average accuracy of 98.10%, outperforming both individual base models and existing ensemble methods. Visualization via Grad-CAM further confirms that the framework focuses on clinically relevant cellular structures, validating its diagnostic relevance. By bridging statistical uncertainty quantification with deep ensemble learning, this work offers a principled and transparent methodology for medical image classification, with potential extensibility to other domains requiring reliable and interpretable predictions under uncertainty. SN - 3070-393X PB - Institute of Central Computation and Knowledge LA - English ER -
@article{Hu2026Quantile,
author = {Hui Hu and Jiewu Huang and Shoumei Cao and Anna Dai},
title = {Quantile Deviation Ensemble Based on Multi-Layer Perceptrons for Cervical Cancer Classification with Uncertainty Perception},
journal = {Journal of Mathematics and Interdisciplinary Applications},
year = {2026},
volume = {2},
number = {1},
pages = {12-27},
doi = {10.62762/JMIA.2026.403659},
url = {https://www.icck.org/article/abs/JMIA.2026.403659},
abstract = {The accurate classification of cervical cytology in Pap smear images remains a critical challenge in computer-aided diagnosis, largely due to the inherent uncertainty and subtle morphological variations among different pathological categories. To address this, we propose a novel uncertainty-aware ensemble framework that integrates statistical quantile analysis with deep learning for robust and interpretable classification. Our framework first leverages three deep convolutional neural networks DenseNet121, MobileNetV2, and ResNet-50 as base feature extractors. Instead of employing naive ensemble strategies, we introduce a quantile deviation based weighting mechanism to dynamically assess and integrate the prediction confidence of each model, explicitly quantifying performance bias across different probability quantiles. This approach not only enhances ensemble stability but also provides a statistical measure of model uncertainty. Subsequently, the weighted probabilistic outputs are fed into a multi-layer perceptron (MLP) for further non-linear optimization and decision refinement, forming a hybrid statistical deep learning pipeline. Evaluated on the publicly available SIPaKMeD dataset, our framework achieves an average accuracy of 98.10\%, outperforming both individual base models and existing ensemble methods. Visualization via Grad-CAM further confirms that the framework focuses on clinically relevant cellular structures, validating its diagnostic relevance. By bridging statistical uncertainty quantification with deep ensemble learning, this work offers a principled and transparent methodology for medical image classification, with potential extensibility to other domains requiring reliable and interpretable predictions under uncertainty.},
keywords = {cervical cytology classification, uncertainty quantification, ensemble learning, quantile deviation, deep learning},
issn = {3070-393X},
publisher = {Institute of Central Computation and Knowledge}
}
 Copyright © 2026 by the Author(s). Published by Institute of Central Computation and Knowledge. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
Copyright © 2026 by the Author(s). Published by Institute of Central Computation and Knowledge. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. Journal of Mathematics and Interdisciplinary Applications
ISSN: 3070-393X (Online)
Email: [email protected]

Portico
All published articles are preserved here permanently:
https://www.portico.org/publishers/icck/