Biomedical Informatics and Smart Healthcare
ISSN: 3068-5524 (Online)
Email: [email protected]


 Submit Manuscript
Edit a Special Issue
Submit Manuscript
Edit a Special Issue
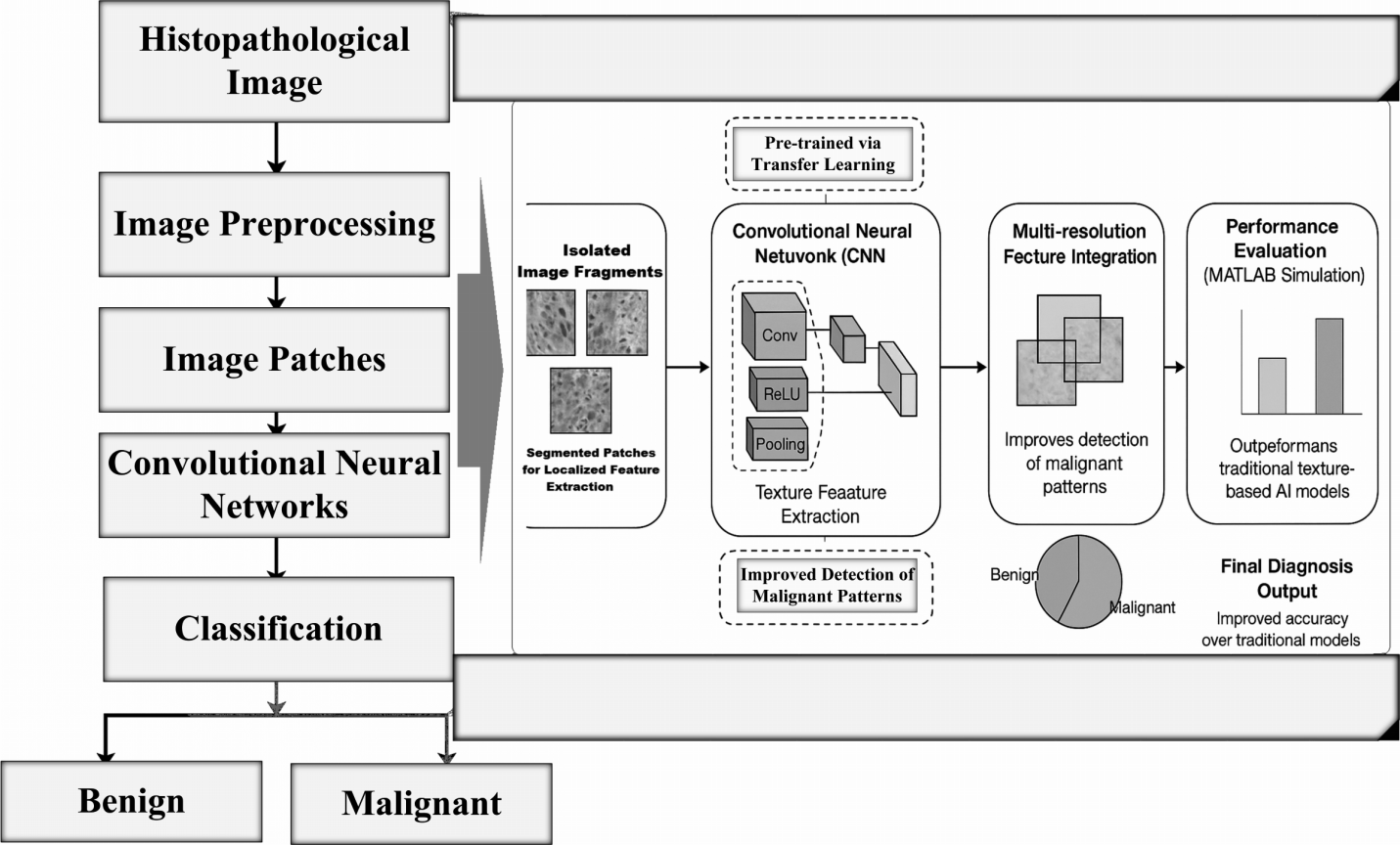
TY - JOUR AU - Hussain, Altaf AU - Hussain, Nasir AU - Wushishi, Usman Jibrin AU - Khalid, Muhammad Imran AU - Wagan, Atif Ali PY - 2025 DA - 2025/11/17 TI - Breast Cancer Image Classification into Benign and Malignant using an Intelligent CNN Framework JO - Biomedical Informatics and Smart Healthcare T2 - Biomedical Informatics and Smart Healthcare JF - Biomedical Informatics and Smart Healthcare VL - 1 IS - 3 SP - 98 EP - 117 DO - 10.62762/BISH.2025.936105 UR - https://www.icck.org/article/abs/BISH.2025.936105 KW - breast cancer detection KW - tissue pathological images KW - benign and malignant images KW - deep learning KW - transfer learning KW - AlexNet KW - CNN AB - Breast cancer is one of the most prevalent and life-threatening diseases among women worldwide. Accurate diagnosis from histopathological biopsy samples is essential, yet manual examination is time-consuming and subject to inter-observer variability, particularly given the shortage of trained pathologists alongside the increasing number of cases. Deep learning, especially Convolutional Neural Networks (CNNs), has emerged as a powerful tool for classifying medical images by automatically extracting discriminative features from raw data. In this study, we investigate the use of the publicly available Breast Cancer Histopathological (BreakHis) image database, which contains benign and malignant samples across multiple magnifications. Our proposed approach extracts isolated image patches, applies CNN-based feature learning and integrates multi-resolution information to improve classification performance. To enhance generalization under limited data, we adopt transfer learning with optimized fine-tuning. Experiments implemented in MATLAB demonstrate that our CNN-based framework achieves higher accuracy than traditional machine learning approaches relying on handcrafted texture descriptors. These findings highlight the potential of CNNs, combined with patch-based multi-resolution analysis, to support pathologists in reliable and efficient breast cancer diagnosis. SN - 3068-5524 PB - Institute of Central Computation and Knowledge LA - English ER -
@article{Hussain2025Breast,
author = {Altaf Hussain and Nasir Hussain and Usman Jibrin Wushishi and Muhammad Imran Khalid and Atif Ali Wagan},
title = {Breast Cancer Image Classification into Benign and Malignant using an Intelligent CNN Framework},
journal = {Biomedical Informatics and Smart Healthcare},
year = {2025},
volume = {1},
number = {3},
pages = {98-117},
doi = {10.62762/BISH.2025.936105},
url = {https://www.icck.org/article/abs/BISH.2025.936105},
abstract = {Breast cancer is one of the most prevalent and life-threatening diseases among women worldwide. Accurate diagnosis from histopathological biopsy samples is essential, yet manual examination is time-consuming and subject to inter-observer variability, particularly given the shortage of trained pathologists alongside the increasing number of cases. Deep learning, especially Convolutional Neural Networks (CNNs), has emerged as a powerful tool for classifying medical images by automatically extracting discriminative features from raw data. In this study, we investigate the use of the publicly available Breast Cancer Histopathological (BreakHis) image database, which contains benign and malignant samples across multiple magnifications. Our proposed approach extracts isolated image patches, applies CNN-based feature learning and integrates multi-resolution information to improve classification performance. To enhance generalization under limited data, we adopt transfer learning with optimized fine-tuning. Experiments implemented in MATLAB demonstrate that our CNN-based framework achieves higher accuracy than traditional machine learning approaches relying on handcrafted texture descriptors. These findings highlight the potential of CNNs, combined with patch-based multi-resolution analysis, to support pathologists in reliable and efficient breast cancer diagnosis.},
keywords = {breast cancer detection, tissue pathological images, benign and malignant images, deep learning, transfer learning, AlexNet, CNN},
issn = {3068-5524},
publisher = {Institute of Central Computation and Knowledge}
}
 Copyright © 2025 by the Author(s). Published by Institute of Central Computation and Knowledge. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
Copyright © 2025 by the Author(s). Published by Institute of Central Computation and Knowledge. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. 
Portico
All published articles are preserved here permanently:
https://www.portico.org/publishers/icck/