Abstract
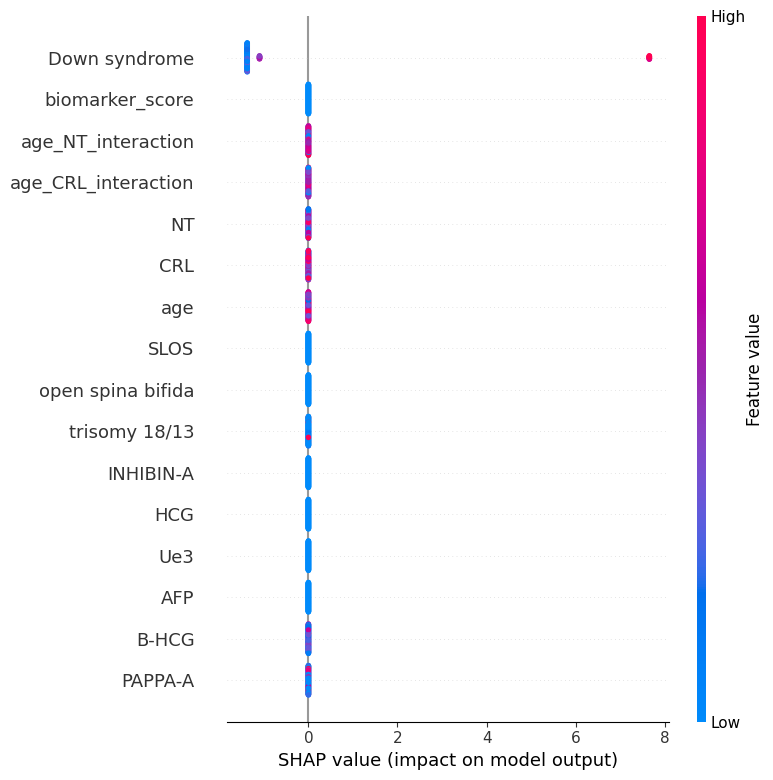
This study developed a machine learning model for early risk stratification of Down syndrome by integrating maternal serum biomarkers and ultrasound measurements. A retrospective multicentre dataset was used, including maternal age, AFP, HCG, INHIBIN-A, and ultrasound parameters (NT, CRL). After imputing missing data and engineering features (e.g., Age_NT_interaction), a Gradient Boosting Machine (GBM) was trained and evaluated using AUROC, precision, recall, and F1-score. The model achieved high performance (AUROC: 0.9921; precision: 1.00; F1-score: 0.91; accuracy: 0.97). SHAP analysis identified key interactions—particularly Age_NT, Age_HCG, and Age_PAPP-A—as major contributors. High maternal age combined with elevated HCG or low PAPP-A was linked to increased risk, aligning with clinical knowledge. The model offers a highly accurate and interpretable approach for Down syndrome risk prediction, supporting personalized, data-driven prenatal care. Prospective validation and clinical integration are recommended.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, Seyed-Ali Sadegh-Zadeh, upon reasonable request.
Funding
This work was supported without any funding.
Conflicts of Interest
The authors declare no conflicts of interest.
Ethical Approval and Consent to Participate
This study was conducted in accordance with the ethical principles and national norms and standards for conducting medical research in Iran, as approved by the Research Ethics Committee of Urmia University of Medical Sciences (Approval ID: IR.UMSU.REC.1403.234, Approval Date: 2024-10-30). Written informed consent was obtained from all participants. The researchers ensured compliance with all professional and legal requirements, maintaining the confidentiality and anonymity of participant data.
Cite This Article
APA Style
Sadegh-Zadeh, S. A., Mamalo, A. S., Saadat, S., Gargari, S. S., Barati, M. A., Mehranfar, S., & Naderi, Z. (2025). Multi-Task Machine Learning for Prenatal Risk Stratification: Integrating Biomarkers, Maternal Age, and Ultrasound Measurements to Predict the Risk of Down Syndrome, Trisomy 18, Trisomy 13, and Neural Tube Defects. Frontiers in Biomedical Signal Processing, 1(1), 24–36. https://doi.org/10.62762/FBSP.2025.954863
Publisher's Note
ICCK stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and Permissions

Copyright © 2025 by the Author(s). Published by Institute of Central Computation and Knowledge. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (
https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.


 Submit Manuscript
Edit a Special Issue
Submit Manuscript
Edit a Special Issue
 Copyright © 2025 by the Author(s). Published by Institute of Central Computation and Knowledge. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
Copyright © 2025 by the Author(s). Published by Institute of Central Computation and Knowledge. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. 