ICCK Journal of Software Engineering
ISSN: 3069-1834 (Online)
Email: [email protected]
 Submit Manuscript
Edit a Special Issue
Submit Manuscript
Edit a Special Issue
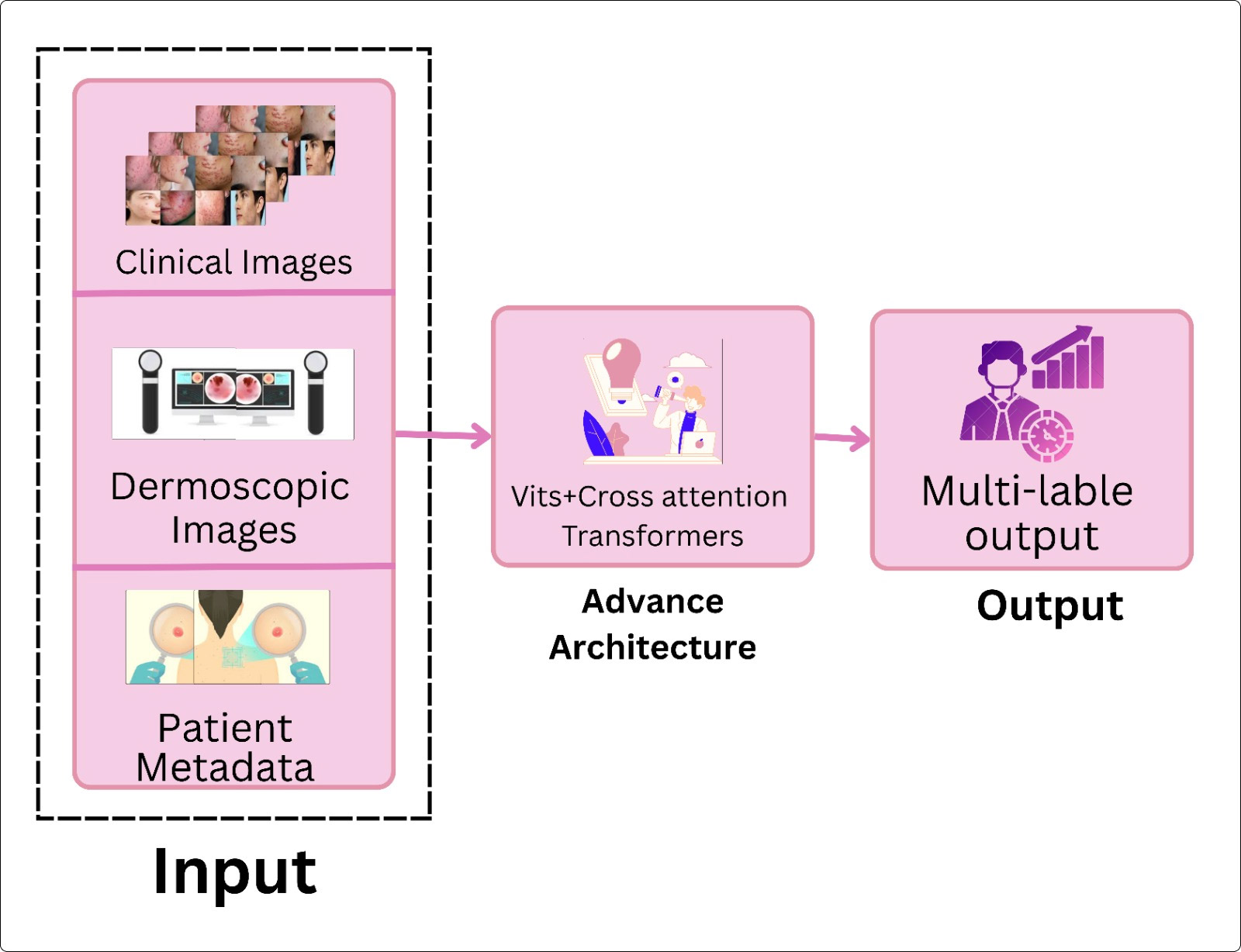
TY - JOUR AU - Nazir, Moomna AU - Ahsan, Azka AU - Khadim, Rabia AU - Abbas, Shakeel AU - Muhammad, Aown AU - Sohail, Zain PY - 2026 DA - 2026/02/11 TI - Software-Engineering Perspectives on Machine for Skin-Disease Classification JO - ICCK Journal of Software Engineering T2 - ICCK Journal of Software Engineering JF - ICCK Journal of Software Engineering VL - 2 IS - 1 SP - 52 EP - 70 DO - 10.62762/JSE.2025.913699 UR - https://www.icck.org/article/abs/JSE.2025.913699 KW - software engineering KW - machine learning KW - deep learning KW - dermatology KW - computer-aided diagnosis KW - MLOps KW - fairness AB - Skin‑disease classification has evolved from simple image recognizers into software‑driven pipelines that demand reliability, reproducibility, and ethical governance. While most AI reviews focus on algorithmic accuracy, few examine these systems through a software‑engineering (SE) lens—essential for assessing pipeline modularity, version control, deployment readiness, and long‑term maintainability, all critical for clinical integration. This review surveys literature from 2015 to early 2025, curating about 180 papers that link skin‑disease classification with SE practices. It traces the shift from handcrafted feature‑based classifiers to end‑to‑end convolutional, ensemble, and transformer architectures, alongside the engineering processes that support versioning, deployment, and monitoring. Benchmark datasets (PH$^2$, HAM10000, ISIC, etc.) have established reproducible evaluation protocols that underpin software verification. Emerging directions—self‑supervised pretraining, multimodal fusion, human‑AI collaboration—signal a move from model‑centric to system‑level integration. The analysis highlights not only accuracy and generalization but also SE quality attributes: scalability, maintainability, explainability, and fairness, which are indispensable for trustworthy adoption in diverse clinical workflows. SN - 3069-1834 PB - Institute of Central Computation and Knowledge LA - English ER -
@article{Nazir2026SoftwareEn,
author = {Moomna Nazir and Azka Ahsan and Rabia Khadim and Shakeel Abbas and Aown Muhammad and Zain Sohail},
title = {Software-Engineering Perspectives on Machine for Skin-Disease Classification},
journal = {ICCK Journal of Software Engineering},
year = {2026},
volume = {2},
number = {1},
pages = {52-70},
doi = {10.62762/JSE.2025.913699},
url = {https://www.icck.org/article/abs/JSE.2025.913699},
abstract = {Skin‑disease classification has evolved from simple image recognizers into software‑driven pipelines that demand reliability, reproducibility, and ethical governance. While most AI reviews focus on algorithmic accuracy, few examine these systems through a software‑engineering (SE) lens—essential for assessing pipeline modularity, version control, deployment readiness, and long‑term maintainability, all critical for clinical integration. This review surveys literature from 2015 to early 2025, curating about 180 papers that link skin‑disease classification with SE practices. It traces the shift from handcrafted feature‑based classifiers to end‑to‑end convolutional, ensemble, and transformer architectures, alongside the engineering processes that support versioning, deployment, and monitoring. Benchmark datasets (PH\$^2\$, HAM10000, ISIC, etc.) have established reproducible evaluation protocols that underpin software verification. Emerging directions—self‑supervised pretraining, multimodal fusion, human‑AI collaboration—signal a move from model‑centric to system‑level integration. The analysis highlights not only accuracy and generalization but also SE quality attributes: scalability, maintainability, explainability, and fairness, which are indispensable for trustworthy adoption in diverse clinical workflows.},
keywords = {software engineering, machine learning, deep learning, dermatology, computer-aided diagnosis, MLOps, fairness},
issn = {3069-1834},
publisher = {Institute of Central Computation and Knowledge}
}
 Copyright © 2026 by the Author(s). Published by Institute of Central Computation and Knowledge. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
Copyright © 2026 by the Author(s). Published by Institute of Central Computation and Knowledge. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. 
Portico
All published articles are preserved here permanently:
https://www.portico.org/publishers/icck/