Next-Generation Computing Systems and Technologies
ISSN: 3070-3328 (Online)
Email: [email protected]
 Submit Manuscript
Edit a Special Issue
Submit Manuscript
Edit a Special Issue
TY - JOUR
AU - Behera, Chandra Shekhar
AU - Bissoyi, Swarupananda
PY - 2025
DA - 2025/12/21
TI - A Framework for Secure and Interoperable Clinical Summarization Using the Model Context Protocol: Integrating MIMIC-III and FHIR with TinyLlama
JO - Next-Generation Computing Systems and Technologies
T2 - Next-Generation Computing Systems and Technologies
JF - Next-Generation Computing Systems and Technologies
VL - 1
IS - 2
SP - 91
EP - 101
DO - 10.62762/NGCST.2025.784852
UR - https://www.icck.org/article/abs/NGCST.2025.784852
KW - model context protocol
KW - tinyLlama
KW - clinical summarization
KW - FHIR
KW - interoperability
KW - MIMIC-III
KW - healthcare AI
KW - data security
KW - ROUGE
KW - BERTScore
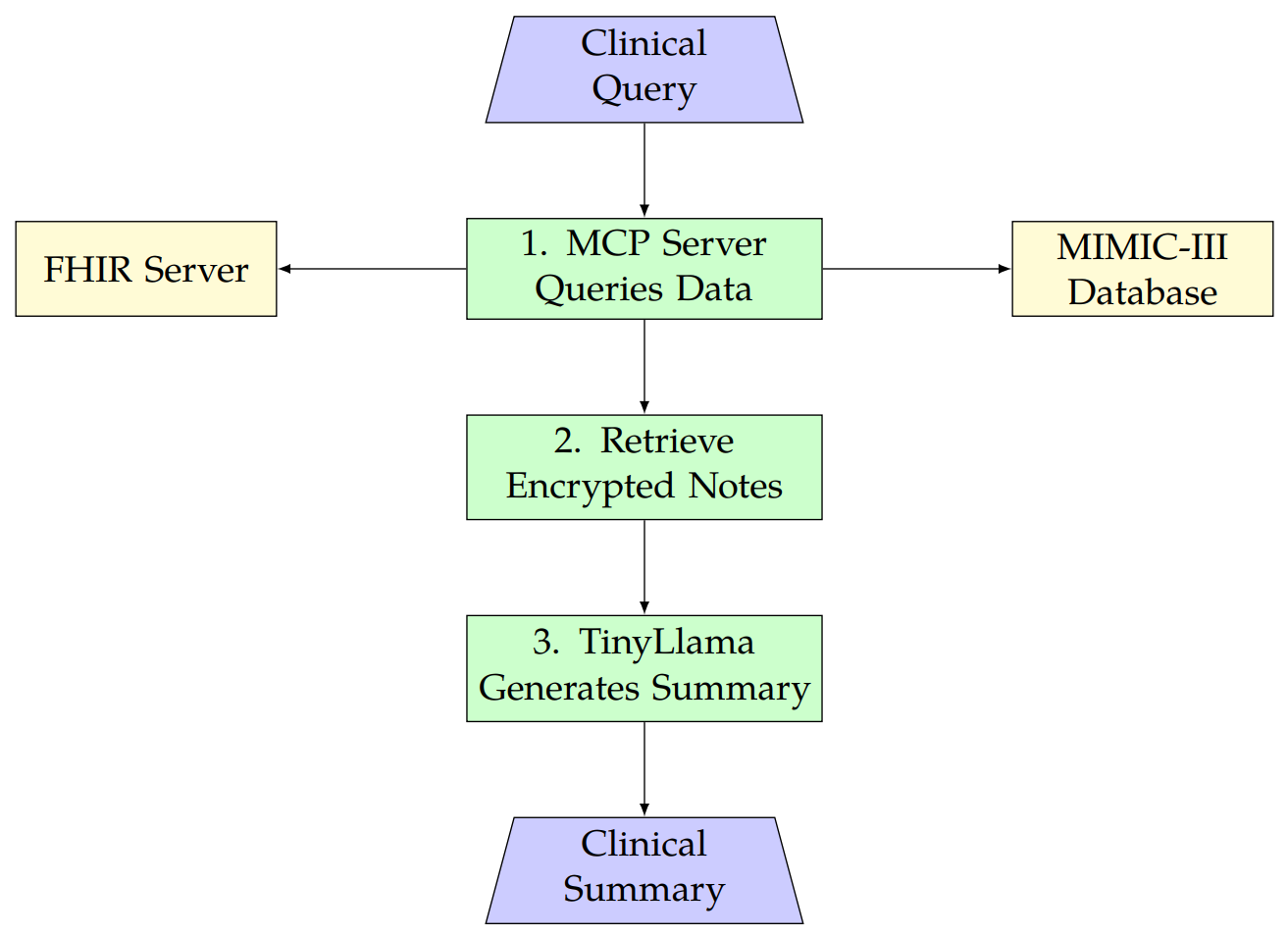
AB - This research presents a new framework for clinical summarization that combines the TinyLlama model with MIMIC-III and FHIR data using the Model Context Protocol (MCP). Unlike cloud-based models like Med-PaLM, our approach uses local processing to cut costs and protect patient data with AES-256 encryption and strict access controls, meeting HIPAA and GDPR standards. It retrieves FHIR-compliant data from public servers (e.g., \texttt{hapi.fhir.org}) for interoperability across hospital systems. Tested on discharge summaries, it achieves ROUGE-L F1 scores of 0.96 for MIMIC-III and 0.84 for FHIR, beating baselines like BioBERT (0.61, p < 0.001) due to efficient preprocessing and MCP’s accurate data grounding. ROUGE, BLEU and BERTScore metrics, along with visualizations, confirm its reliability. The entire pipeline code is available at \url{https://github.com/shekhar-ai99/clinical-mcp} for transparency and reproducibility.
SN - 3070-3328
PB - Institute of Central Computation and Knowledge
LA - English
ER -
@article{Behera2025A,
author = {Chandra Shekhar Behera and Swarupananda Bissoyi},
title = {A Framework for Secure and Interoperable Clinical Summarization Using the Model Context Protocol: Integrating MIMIC-III and FHIR with TinyLlama},
journal = {Next-Generation Computing Systems and Technologies},
year = {2025},
volume = {1},
number = {2},
pages = {91-101},
doi = {10.62762/NGCST.2025.784852},
url = {https://www.icck.org/article/abs/NGCST.2025.784852},
abstract = {This research presents a new framework for clinical summarization that combines the TinyLlama model with MIMIC-III and FHIR data using the Model Context Protocol (MCP). Unlike cloud-based models like Med-PaLM, our approach uses local processing to cut costs and protect patient data with AES-256 encryption and strict access controls, meeting HIPAA and GDPR standards. It retrieves FHIR-compliant data from public servers (e.g., \texttt{hapi.fhir.org}) for interoperability across hospital systems. Tested on discharge summaries, it achieves ROUGE-L F1 scores of 0.96 for MIMIC-III and 0.84 for FHIR, beating baselines like BioBERT (0.61, p < 0.001) due to efficient preprocessing and MCP’s accurate data grounding. ROUGE, BLEU and BERTScore metrics, along with visualizations, confirm its reliability. The entire pipeline code is available at \url{https://github.com/shekhar-ai99/clinical-mcp} for transparency and reproducibility.},
keywords = {model context protocol, tinyLlama, clinical summarization, FHIR, interoperability, MIMIC-III, healthcare AI, data security, ROUGE, BERTScore},
issn = {3070-3328},
publisher = {Institute of Central Computation and Knowledge}
}
 Copyright © 2025 by the Author(s). Published by Institute of Central Computation and Knowledge. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
Copyright © 2025 by the Author(s). Published by Institute of Central Computation and Knowledge. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. Next-Generation Computing Systems and Technologies
ISSN: 3070-3328 (Online)
Email: [email protected]

Portico
All published articles are preserved here permanently:
https://www.portico.org/publishers/icck/